Malaiyalam Mariappan
Associate Professor

Ph.D. in Biochemistry
University of Göttingen, Germany
Department of Cell Biology and Biochemistry
Texas Tech University Health Sciences Center
3601 4th Street Lubbock, TX 79430-6540
Office Phone: (806) 743-4104
Malaiyalam.Mariappan@ttuhsc.edu
Research Interests
Proteins carry out most cellular functions and are responsible for the structure, function, and regulation of all organisms on Earth. Yet, we do not fully understand how proteins are properly produced and degraded (trashed) when they become defective in mammalian cells. Failure in any of these processes can lead to devastating human diseases, including type 2 diabetes, cystic fibrosis, Alzheimer's, and Parkinson's disease. Mariappan's lab has three projects that aim to understand the protein life cycle using biochemical, fluorescence imaging, CRISPR/Cas9 editing, proteomics, and structural approaches.
Current Projects
Project 1: Membrane Protein Biogenesis and Quality Control
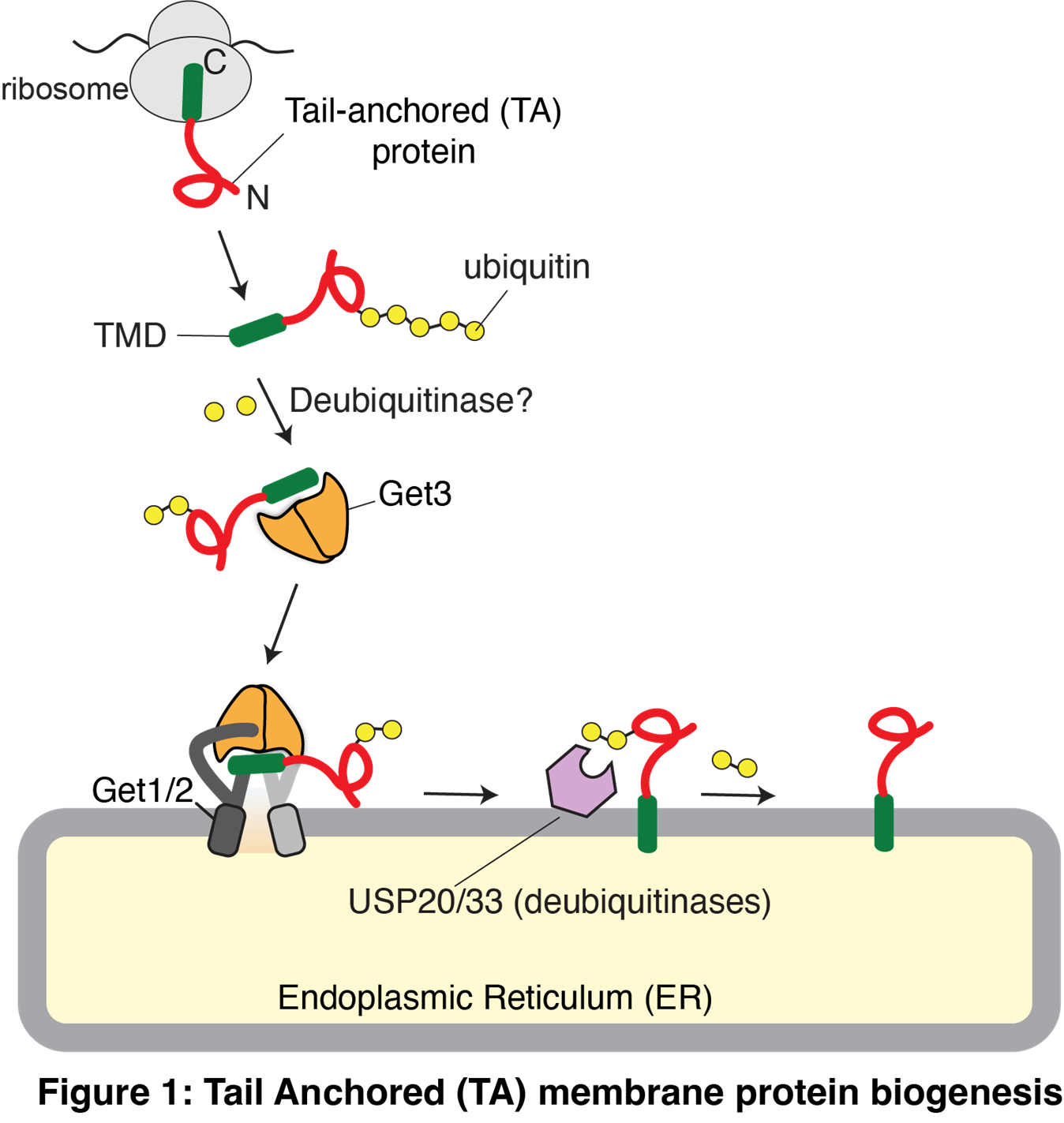
About 30% of the human proteome consists of membrane proteins, which are crucial for cell physiology, including cell-cell communication, signal transduction, and molecular transport across membranes. Due to their high hydrophobicity, membrane proteins encounter challenges during biogenesis and degradation, which are further exacerbated for tail-anchored (TA) proteins, a vital class of membrane proteins. TA proteins contain a single C-terminal transmembrane domain (TMD) (Figure 1). They are post-translationally targeted and inserted into the membranes of the endoplasmic reticulum (ER), mitochondria, or peroxisomes. Our research, along with others, has identified numerous factors mediating the targeting and insertion of TA proteins into the ER membrane through the GET (Guided Entry of Tail-anchored proteins) pathway (Mariappan et al., Nature 2010; Mariappan et al., Nature 2011; Heo et al., Cell Reports 2023)
We recently discovered that newly synthesized TA proteins are polyubiquitinated in cells. Polyubiquitinated proteins, particularly those decorated with K48-linked ubiquitin chains, are typically routed to proteasomal degradation. Instead, polyubiquitinated TA proteins are properly captured by the targeting chaperone GET3 (TRC40) and delivered to the ER membrane, where the USP20/33 deubiquitinases subsequently remove their ubiquitin modifications (Culver and Mariappan Journal of Cell Biology, 2021). Our ongoing studies aim to understand why TA proteins are initially decorated with ubiquitin chains and why these modifications are later removed. What is the role of ubiquitin modification in TA protein insertion and degradation? Addressing these questions will likely explain why mutations in GET3 are associated with Parkinson's disease in humans.
Project 2: ER Stress and the Unfolded Protein Response (UPR)
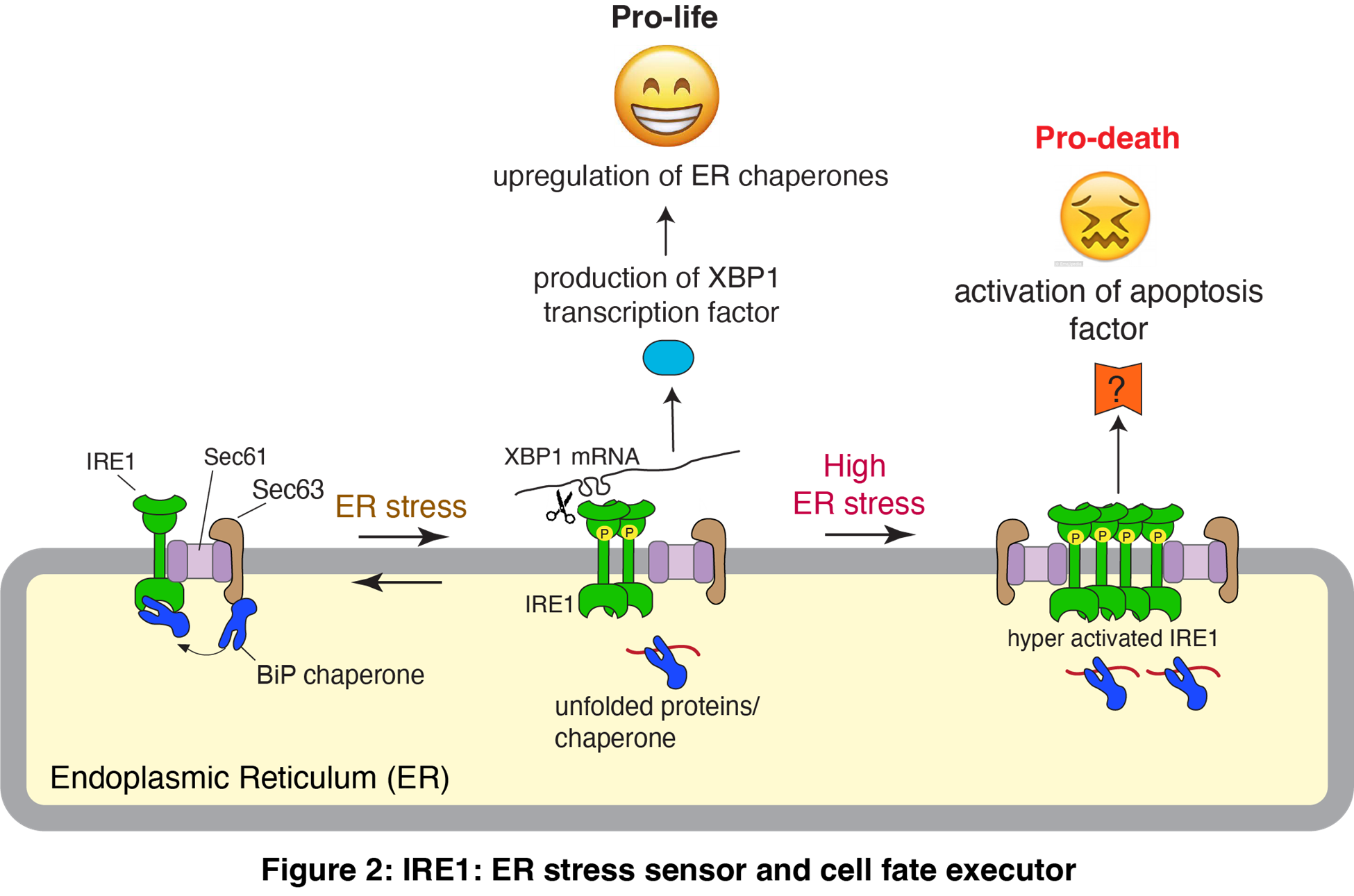
The endoplasmic reticulum (ER) is crucial for synthesizing secretory and membrane proteins, including antibodies, growth factors, and receptors. However, its protein synthesis often surpasses its folding capacity, resulting in the accumulation of misfolded proteins and causing ER stress. The unfolded protein response (UPR) plays a significant role in adjusting the protein folding capacity of the ER to the incoming protein load. IRE1 is a conserved UPR sensor that detects misfolded proteins in the ER lumen and activates the transcription factor XBP1 to alleviate ER stress (Figure 2). If ER stress is not mitigated, IRE1 can also mediate cell death by less understood mechanisms, which is implicated in type 2 diabetes and neurodegenerative diseases. Therefore, it is essential to understand how IRE1 activity is regulated in human cells to develop better strategies to treat these diseases.
Studies from my laboratory discovered that IRE1 exists in a direct complex with the Sec61 protein translocation channel, to which the SRP pathway recruits its substrate XBP1 mRNA (Plumb et al., eLIFE 2015). We have further shown that the Sec61 translocon-associated protein Sec63 recruits the luminal chaperone BiP to bind onto IRE1, thus turning off IRE1 signaling once ER stress is alleviated (Sundaram et al., eLIFE 2017; Li et al., Cell Reports 2020). Without the Sec complex, IRE1 is hyperactivated and induces cell death in insulin-secreting pancreatic beta cells, which is implicated in type 2 diabetes. Our current studies focus on understanding how IRE1 makes life-or-death decisions during ER stress in neuronal and pancreatic beta cells (Figure 2). Additionally, we aim to obtain the structural information of the IRE1/Sec61/Sec63 complex to understand how IRE1 senses the accumulation of unfolded proteins in the ER.
Project 3: The ER-Associated Protein Degradation Pathway
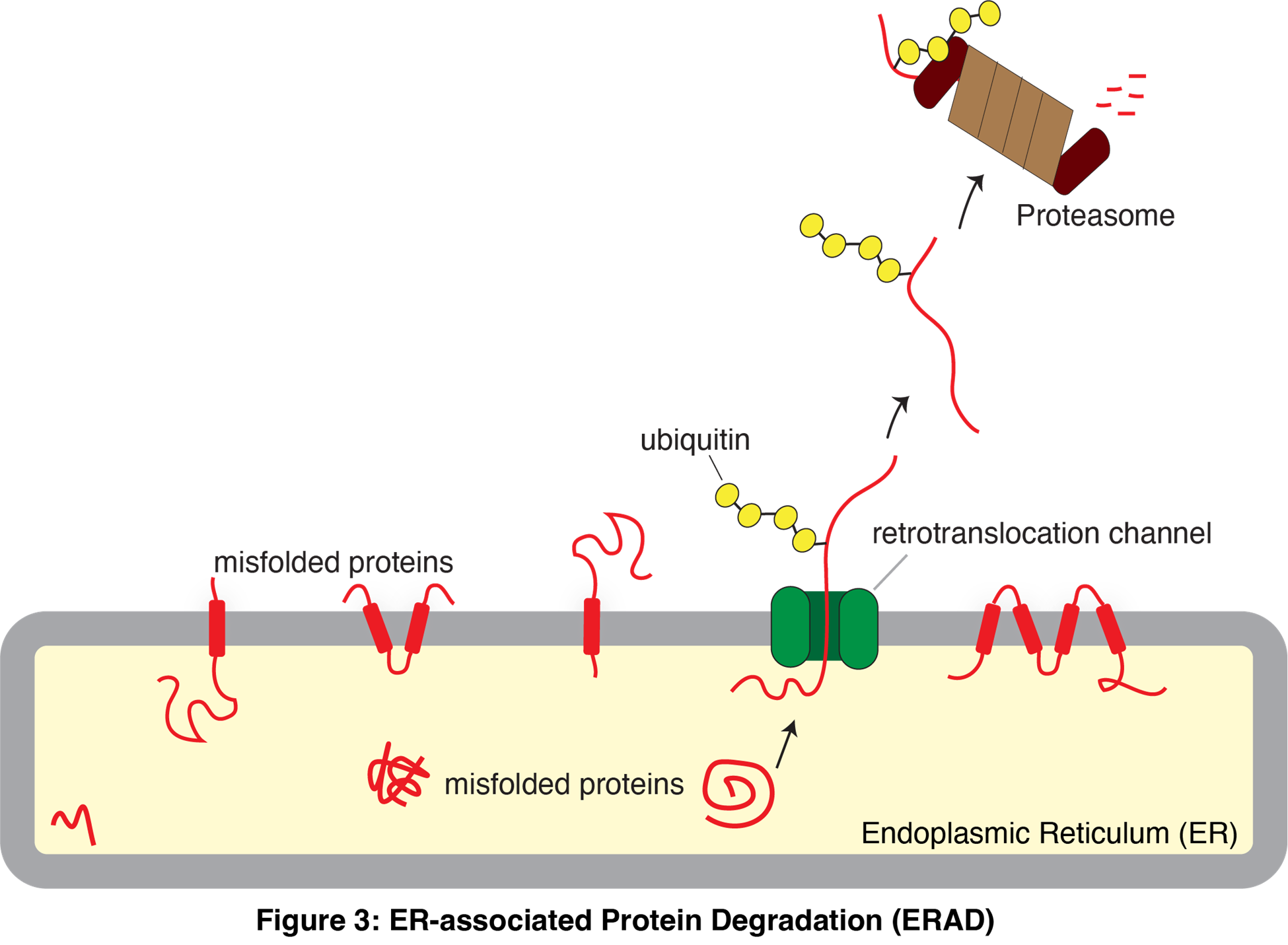
While recent studies mapped out protein quality control pathways in various cellular organelles, we still need to fully understand how these pathways identify the many misfolded substrates, each differing in size, shape, and structure (Figure 3). The endoplasmic reticulum (ER) is a major site where misfolded proteins are generated because it produces complex proteomes with intricate topologies and many co- and post-translational modifications. Misfolded proteins in the ER are usually retrotranslocated to the cytosol and targeted for degradation by the proteasome through multiple ER-associated degradation (ERAD) pathways.
Although 25 different membrane-bound E3 ligases mediate the ubiquitination of misfolded proteins in the ER, much remains unknown about their identities and physiological roles. To address this, we are developing novel proteomic strategies combined with CRISPR/Cas9 technology to identify the endogenous substrates of ERAD pathways. These approaches will help reveal the molecular mechanisms by which E3 ligases recognize substrates with diverse structures in the ER. Additionally, these proteomic tools are expected to be valuable in identifying proteins prone to misfolding during stress, aging, and viral infection.
Undergraduate students, graduate students, and postdoctoral fellows interested in any of these projects should email: malaiyalam.mariappan@ttuhsc.edu.
Selected Publications:
- Mariappan M, Li X, Stefanovic S, Sharma A, Mateja A, Keenan RJ, Hegde RS. A ribosome-associating factor chaperones tail-anchored membrane proteins. Nature. 2010, 466(7310):1120-4.
- Hessa T, Sharma A, Mariappan M, Eshleman HD, Gutierrez E, Hegde RS. Protein targeting and degradation are coupled for elimination of mislocalized proteins. Nature. 2011, 475(7356):394-7.
- Mariappan M*, Mateja A*, Dobosz M, Bove E, Hegde RS, Keenan RJ. The mechanism of membrane-associated steps in tail-anchored protein insertion. Nature. 2011, 477(7362):61-6 (*shared first authorship).
- Plumb R*, Zhang ZR*, Appathurai S, Mariappan M. A functional link between the co-translational protein translocation pathway and the UPR. eLIFE. 2015 May 20;4:e07426. PMID: 25993558; PMCID: PMC4456659 (*shared first authorship).
- Sundaram A*, Plumb R*, Appathurai S, Mariappan M. The Sec61 translocon limits IRE1α signaling during the unfolded protein response. eLIFE. 2017 May 15;6:e27187. PMID: 28504640; PMCID: PMC5449187 (*shared first authorship).
- Sundaram A, Appathurai S, Plumb R, Mariappan M. Dynamic changes in complexes of IRE1α, PERK, and ATF6α during endoplasmic reticulum stress. Mol Biol Cell. 2018 Jun 1;29(11):1376-1388. PMID: 29851562; PMCID: PMC5994896.
- Sun S, Mariappan M. Lonely ER Membrane Proteins Travel to the Nucleus to Rest in Peace by the Asi Complex. Mol Cell. 2020 Jan 2;77(1):1-2. PMID: 31951515.
- Culver JA, Mariappan M. Membrane Protein Biogenesis: PAT Complex Pats Membrane Proteins into Shape. Current Biology. 2020 Nov 16;30(22):R1387-R1389. PMID: 33202243.
- Li X, Sun S, Appathurai S, Sundaram A, Plumb R, Mariappan M. A Molecular Mechanism for Turning Off IRE1α Signaling during Endoplasmic Reticulum Stress. Cell Reports 2020 Dec 29;33(13):108563. PMID: 33378667; PMCID: PMC7809255.
- Culver JA, Mariappan M. Deubiquitinases USP20/33 promote the biogenesis of tail-anchored membrane proteins. Journal of Cell Biology. 2021 May 3;220(5):e202004086. PMID: 33792613; PMCID: PMC8020466.
- Sun S, Li X, Mariappan M. Signal sequences encode information for protein folding in the endoplasmic reticulum. Journal of Cell Biology. 2023 Jan 2;222(1):e202203070. Epub 2022 Dec 2. PMID: 36459117; PMCID: PMC9723807.
- Heo P#, Culver JA, Miao J, Pincet F#, Mariappan M#. The Get1/2 insertase forms a channel to mediate the insertion of tail-anchored proteins into the ER. Cell Reports. 2023 Jan 31;42(1):111921. Epub 2022 Dec 28. PMID: 36640319; PMCID: PMC9932932 (# shared corresponding authorship).
- Guay KP, Ke H, Canniff NP, George GT, Eyles SJ, Mariappan M, Contessa JN, Gershenson A, Gierasch LM, Hebert DN. ER chaperones use a protein folding and quality control glyco-code. Molecular Cell. 2023, S1097-2765(23)00922-X. PMID: 38052210.
- Li X. and Mariappan M. Nascent Chain Ubiquitination is Uncoupled from Degradation to Enable Protein Maturation. bioRxiv [Preprint]. 2023 Oct 10:2023.10.09.561585. doi: org/10.1101/2023.10.09.561585. PMID: 37873109; PMCID: PMC10592752 (in revision).
